

Protein interaction module that occurs in humans, responsible for the glutamatergic receptor signaling pathway. The genes responsible for producing 14 proteins found in the network (nodes in green) were identified by the tool as being associated with schizophrenia
Concepts from graph theory and complex networks together with tools such as gene network inference help map structural and functional aspects between genes, pinpointing therapeutic targets for diseases.
Concepts from graph theory and complex networks together with tools such as gene network inference help map structural and functional aspects between genes, pinpointing therapeutic targets for diseases.
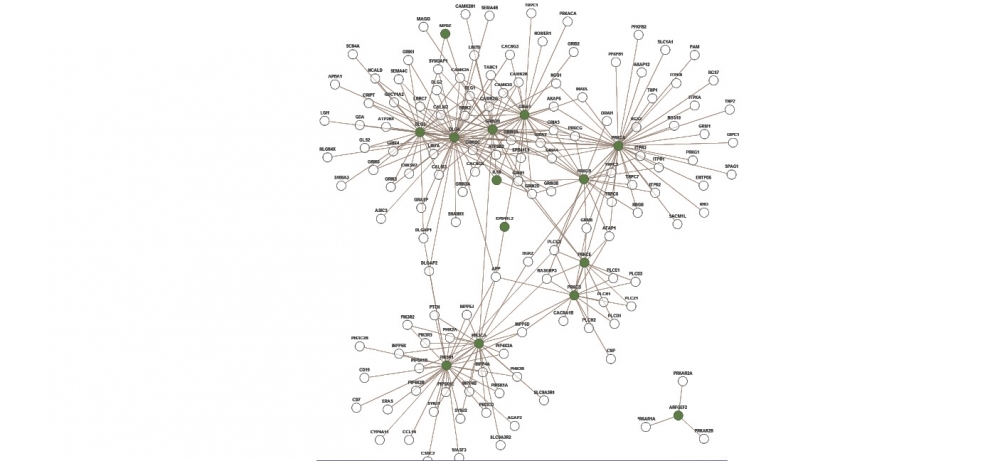
Protein interaction module that occurs in humans, responsible for the glutamatergic receptor signaling pathway. The genes responsible for producing 14 proteins found in the network (nodes in green) were identified by the tool as being associated with schizophrenia
By Karina Toledo, in Ohio | Agência FAPESP – Bioinformatic tools have been used by researchers at the Federal University of the ABC (UFABC) and the University of São Paulo (USP) to model genetic interaction networks, reveal functional relationships and thus identify targets for the treatment of complex diseases such as schizophrenia, autism and cancer.
Advances in this field were presented by David Corrêa Martins Junior, a professor from the Center for Mathematics, Computation and Cognition (CMCC) of the UFABC on March 31 in Columbus, Ohio during FAPESP Week Michigan-Ohio. The event was designed to promote increased collaboration among researchers from the U.S. and São Paulo.
Some of the findings have also been published in articles in the journals Information Sciences and BMC Bioinformatics.
"A single complex disease can manifest itself in many different ways in each individual, with varying degrees of severity. The goal of this type of study is to be able to offer individualized treatment, taking into account these particularities,” Martins Junior explained in an interview with Agência FAPESP.
The studies are based both on clinical data – gene expression, interaction among proteins and DNA methylation – held in public databases as well as data from patients under the care of collaborators such as Professor Helena Brentani of the Psychiatry Institute at the University of São Paulo (IPq-USP) who works with carriers of autism.
Concepts from graph theory and complex networks, together with bioinformatics tools, are enabling scientists to compare gene expression and centrality in the gene networks between carriers of a particular disease and individuals in a control group. It is also possible to compare data from patients with the same disease but different degrees of severity, or even to evaluate how gene expression and the structure of a gene network in one person changes in different contexts and at different times. The idea is to try to discover how the genes are connected and how this circuit controls the various cell functions.
"When we don’t know the function of a gene, we can try to compare its expression or the structure of its vicinity in the gene network with others that present a similar signal and whose function is known, and thus formulate a hypothesis. If we have a target gene and we want to know to what extent its expression depends on the signal from other genes, we can use tools like gene network inference or gene prioritization," Martins Junior explained.
According to the UFABC researcher, genes that have already been associated with diseases in previous studies can serve as a starting point for analyses. "We can evaluate which of the other genes are most closely associated with them in terms of expression and in terms of connective structure. Most likely, they will also be involved in the disease in question,” he said.
In a post-doctoral research study conducted by Sergio Nery Simões during his graduate program in bioinformatics at USP, a tool known as NERI (Network Medicine Relative Importance) was developed that integrates gene expression data, protein integration networks and data from association studies (genes known to be associated with a particular disease) to identify new genes that may also be associated with the illness in question.
"We did a case study using public databases on schizophrenia and saw that many of the genes obtained by the NERI tool were associated with the so-called glutamatergic pathway, responsible for the transmission and reception of neurotransmitters during neuronal synapse. This pathway is well-cited in the literature as being associated with the disease. This suggests that other genes identified by the tool also have the potential of being related to schizophrenia,” Martins Junior explained.
The group’s research is funded by FAPESP through two thematic projects – one coordinated by João Eduardo Ferreira at the Institute of Mathematics and Statistics (IME) at USP and the other by Roberto Marcondes Cesar Junior also at the IME-USP. There is also a regular research grant coordinated by Raphael Yokoingawa de Camargo at the CMCC-UFABC.
Analysis of social networks
On the same panel dedicated to "Bioinformatics and Data Analytics,” OSU Computer Science and Engineering Professor Srinivasan Parthasarathy talked about how information collected on social networks such as Twitter or WhatsApp can be used to predict or mitigate problems related to natural disasters and epidemics.
Working in partnership with researchers from several countries, including Brazilians associated with the Federal University of Minas Gerais (UFMG), Parthasarathy is gathering and analyzing information from short messages sent during extreme climatic events such as hurricanes and floods or during epidemic outbreaks and using the contents to feed computer models. The goal is to identify problems before they become critical and provide information that can help teams that render assistance to the site to plan control strategies.
"If I have 5,000 people available to provide assistance, I have to identify the areas most affected that need to be prioritized. To that end, we use the information collected from social media together with the physical data that traditionally feeds the hurricane and flood prevention models, such as wind and atmospheric pressure, which come from a more traditional form of sensing,” the researcher explained.
The greatest challenge, according to Parthasarathy, is the ability to filter messages that are actually related to the problem in question. “There is a lot of noise on social networks. For example, if our objective is monitoring a dengue epidemic in a particular city, we have to be able to isolate the tweets that originated in that area and include that information in our model. By integrating social and physical sensors, we can produce better results,” said the researcher.
Also taking part in the panel convened during FAPESP Week were Álvaro Montenegro, a professor in the Department of Geography at Ohio State University, and João Luiz Filgueiras de Azevedo of the Aeronautics and Space Institute (IAE).
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





