
Parameter found by Brazilian, Chinese and American researchers indicates the efficiency of a protein folding into its functional shape
Parameter found by Brazilian, Chinese and American researchers indicates the efficiency of a protein folding into its functional shape
Parameter found by Brazilian, Chinese and American researchers indicates the efficiency of a protein folding into its functional shape

Parameter found by Brazilian, Chinese and American researchers indicates the efficiency of a protein folding into its functional shape
By Karina Toledo
Agência FAPESP – For a protein to perform its function inside an organism—whether it is structural, enzymatic, hormonal, energetic, defensive or transporting nutrients—the amino acid chain that composes it must form itself into a specific three-dimensional shape.
For decades, scientists have been trying to discover how this very complex process, known as protein folding, happens in a matter of seconds or even thousandths of a second. A FAPESP-funded study published in the Proceedings of the National Academy of Sciences (PNAS) put a few pieces of the jigsaw puzzle in place.
“Studies carried out in the 1960s showed that if proteins had to randomly explore all possible configurations until they reached their native conformation, the folding process would last for a time period equivalent to the age of the universe,” said Vitor Barbanti Pereira Leite, advisor for the doctoral studies of Ronaldo J. Oliveira, who was the first author on the article.
It was already known that all the information necessary for folding to occur was contained in the amino acid sequence after it was reproduced in test tubes without the influence of biological factors. The hypothesis then arose that there could be a route that facilitates arrival at the functional state.
“Twenty years passed while scientists tried to uncover the possible routes and identify the intermediate stages until they realized that this wasn’t the mechanism,” said Leite.
In the 1990s, José Nelson Onuchic, a Brazilian professor at Rice University in Houston and one of the co-authors of the recently published PNAS article, introduced the idea that there is not a single path but rather a mechanism in which all the intermediate stages lead to the native structure.
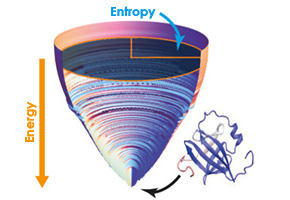
Because the protein would have a certain energy in each of the alternative configurations, Onuchic proposed that if the energy at each intermediate stage were mapped out, this relief—known as the energy landscape—would have a funnel shape. The native structure would be at the bottom of the funnel, which represents the most stable energy state.
“Imagine a blind person trying to randomly hit a ball into a hole on a golf course. If the course were flat, he would take his entire lifetime to find it. However, if the course were funnel shaped, the ball would roll to the center no matter where it was hit,” explained Leite.
With the help of simplified computer models, Leite and his team were able, for the first time, to measure the dimensions of the energy funnel—reinforcing the theory proposed by Onuchic. Then, based on the funnel measurements, they developed a parameter—called the landscape describer (Λ)—that was able to indicate the efficiency of each protein’s folding process.
“To calculate Λ, we use three different measurements: the roughness of the energy landscape and the width and depth of the funnel,” said Leite.
The roughness is calculated by the variation in energy that occurs when the protein jumps from one configuration to the next. “The rougher the landscape, the more difficult it will be to get to the bottom of the funnel,” he said.
The width varies according to the number of possible configurations that are accessed by the protein. “The greater the entropy of the folding state, meaning the number of possible combinations when the protein is unfolded, the larger the mouth of the funnel will be,” Leite explained.
Last, the depth represents the distance in energy between the native conformation and the completely unfolded state. “We measured the decrease in energy when the protein moves from the denatured state to the functional state,” he said.
Proteins have been selected during the evolutionary process to work at physiological temperatures—approximately 36° Celsius in humans.
“Temperature directly influences the entire system. If we heat certain proteins to a little above the physiological temperature, they begin to unfold. The energy landscape remains the same, but the folded state is no longer as stable,” explained Leite.
The researchers studied a group of 21 proteins of different shapes and sizes and showed that the Λ parameter strongly correlates with stability and the time a protein takes to fold.
“The stability and folding time were calculated by simulation, but they correlated with experimental data from the scientific literature,” said Leite.
The bridge between theory and practice
The study was developed on the São José do Rio Preto campus of the Universidade Estadual Paulista (Unesp). Researchers from Rice University in Houston and the Chinese Academy of Sciences collaborated on the work.
In addition to creating a bridge between theoretic and experimental results, the study increased the understanding of the protein-folding process, which according to Leite, could be useful to researchers in many fields, especially health.
“Many illnesses, like Alzheimer’s, Parkinson’s, cystic fibrosis, phenylketonuria and cancer, are related to poor protein function. In these pathologies, there is no external agent. Rather, the organism itself cannot maintain the proteins in their native state for some reason,” said Leite.
Knowledge of the folding process can also be applied in fields like bioenergy. Currently, Leite is coordinating studies with the Brazilian Bioethanol Science and Technology Laboratory (CTBE) in the hope of developing enzymes for bioethanol manufacturing.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





