
A mechanism helps prevent protein aggregation inside cells, which can cause illnesses such as Alzheimer’s and Parkinson’s
The presence of oxygen and the development of aerobic metabolism on Earth allowed living beings to take advantage of the energy in foods in a much more efficient manner. This evolutionary achievement, however, came at a price: it left cells vulnerable to the effects of oxidizing substances.
The presence of oxygen and the development of aerobic metabolism on Earth allowed living beings to take advantage of the energy in foods in a much more efficient manner. This evolutionary achievement, however, came at a price: it left cells vulnerable to the effects of oxidizing substances.

A mechanism helps prevent protein aggregation inside cells, which can cause illnesses such as Alzheimer’s and Parkinson’s
By Karina Toledo
Agência FAPESP – The presence of oxygen and the development of aerobic metabolism on Earth allowed living beings to take advantage of the energy in foods in a much more efficient manner. This evolutionary achievement, however, came at a price: it left cells vulnerable to the effects of oxidizing substances.
These byproducts of aerobic respiration interact with proteins, lipids, carbohydrates and nucleic acids, disrupting the functions of these macromolecules. This process can lead to cell death, and in more complex organisms such as humans, it is thought to be one basis for diseases like cancer, arthritis, atherosclerosis, Parkinson’s and Alzheimer’s.
Fortunately, most organisms have developed mechanisms to protect themselves against damage from oxidation. One such mechanism was recently discovered by Brazilian researchers and was featured on the cover of Antioxidants & Redox Signaling, one of the most prominent journals in the field.
The study, financed by FAPESP under its Regular Research Support program and coordinated by Marilene Demasi at the Instituto Butantan’s Biochemistry and Biophysics Laboratory, revealed the strategy used by the yeast Saccharomyces cerevisiae to accelerate the breakdown of oxidized proteins.
“Aside from losing their function, proteins damaged by oxidation tend to aggregate, and today we know that this is the cause of a number of neuropathologies. The best defense for cells is to degrade these molecules,” explained Demasi.
Freeing cells of undesired proteins, whether oxidized or not, is the mission of a protein complex called the proteasome. “The proteasome regulates many different functions, such as responses to internal and external stimuli, cell division and cell death. This regulation is carried out by degrading proteins that are involved in these processes,” explained Demasi.
This system has been maintained throughout evolution in all eukaryotes (organisms whose cells have a nucleus isolated from the cytoplasm by a membrane and possess many organelles). It is present, therefore, in many unicellular organisms as well as in multicellular plants, fungi and animals.
“We knew that under oxidative stress, the proteasome goes through a process called glutathiolation, and we wanted to understand why. Our study showed for the first time that the glutathiolated proteasome is able to degrade oxidized proteins at a faster rate and at a lower energetic cost to the cell,” said Demasi.
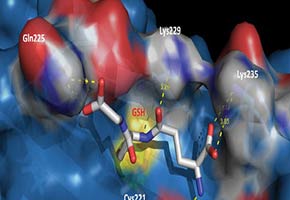
Glutathiolation, she explained, is an oxidizing modification that affects the residues of cysteine amino acids present in the proteasome. “However, this is a non-deleterious and reversible oxidizing modification that works as a cell protection mechanism,” she affirmed.
Opening the gates
Proteins that need to be eliminated during normal cell regulation processes are marked with another protein, called ubiquitin, which targets them for elimination by the proteasome. Scientists know, however, that oxidized proteins can be degraded without ubiquitination.
In order to understand precisely what happens inside the proteasome, the researchers turned to transmission electron microscopy and a technique known as small-angle X-ray scattering (SAXS), developed by Cristiano de Oliveira’s team from the USP Physics Institute. This method allows the analysis of the molecule in a solution and measurements from structural modeling.
“The proteasome has a cylindrical structure, with openings at both ends. However, these openings normally stay closed. We managed to show that when the proteasome is glutathiolated, the gates open, allowing the oxidized protein to enter,” said Demasi.
When the researchers used mass spectrometry, they discovered that only two of the 32 cysteines present in the proteasome undergo glutathiolation—and those two cysteines are related to the opening and closing of the catalytic chamber, where the proteins are degraded. This portion of the work was carried out in collaboration with the team of Professor Fabio Gozzo from the Universidade Estadual de Campinas (UNICAMP).
“One of the glutathiolated cysteines we found is highly conserved along the whole evolutionary chain, from yeast to humans,” said Demasi. “This is a very important result because no one had shown that the proteasome undergoes redox regulation before.”
The study was part of a Thematic Project coordinated by biologist Luis Eduardo Soares Netto of the USP Biosciences Institute.
Demasi and Netto are also connected to the National Institute of Science and Technology (INCT) of Redox Processes in Biomedicine (Redoxome).
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





