

Brazilians participate in international project to boost capacity of PETase to break down polyethylene terephthalate (PET), used in bottles and responsible for producing millions of tons of waste (PETase is shown in blue, with PET chain (yellow) bound to active site, where it will be degraded / image: Rodrigo Leandro Silveira)
Brazilians participate in international project to boost capacity of PETase to break down polyethylene terephthalate (PET), used in bottles and responsible for producing millions of tons of waste.
Brazilians participate in international project to boost capacity of PETase to break down polyethylene terephthalate (PET), used in bottles and responsible for producing millions of tons of waste.
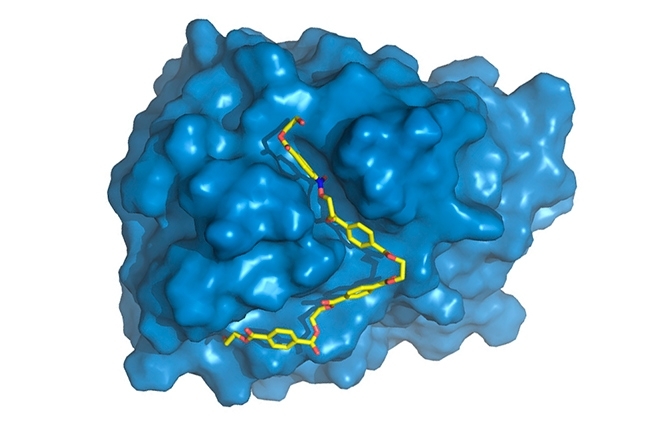
Brazilians participate in international project to boost capacity of PETase to break down polyethylene terephthalate (PET), used in bottles and responsible for producing millions of tons of waste (PETase is shown in blue, with PET chain (yellow) bound to active site, where it will be degraded / image: Rodrigo Leandro Silveira)
By José Tadeu Arantes | Agência FAPESP – Between 4.8 billion and 12.7 billion kilograms of plastic are dumped in the oceans every year, and if this trend continues, the volume will increase tenfold by 2025, according to a study based on data for 2010 and published in the journal Science in 2015.
One of the properties of plastic that explains why it is so widely used is also the main reason it is so harmful to the environment: its resistance to degradation. Bottles and other objects made of PET (polyethylene terephthalate) take at least 800 years to biodegrade in landfills or the sea.
In this context, it is easy to understand the strong interest aroused by the discovery of an enzyme that can digest PET. Now, that enzyme, known as PETase, has had its capacity to break down plastic boosted. The innovation is described in an article published in Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Two researchers affiliated with the University of Campinas’s Chemistry Institute (IQ-UNICAMP) in Brazil collaborated with scientists at the University of Portsmouth in the United Kingdom and the US National Renewable Energy Laboratory. They are postdoctoral fellow Rodrigo Leandro Silveira and his supervisor, Munir Salomão Skaf, Full Professor and Pro-Rector for Research at UNICAMP.
Their participation was supported by FAPESP via a Postdoctoral Scholarship and a Scholarship for a Research Internship Abroad awarded to Silveira and via the Center for Computational Engineering and Sciences (CCES), one of the Research, Innovation and Dissemination Centers (RIDCs) funded by FAPESP. Skaf is the Director of CCES.
“Used mainly to make drink bottles, PET is also widely used in clothing, carpets and other objects. In our research project, we characterized the three-dimensional structure of the enzyme that can digest this plastic, engineered it to boost its degradation capacity, and demonstrated that it also acts on polyethylene-2,5-furandicarboxylate (PEF), a PET substitute made from renewable raw materials,” Silveira told Agência FAPESP.
Interest in PETase arose in 2016, when a group of Japanese researchers led by Shosuke Yoshida identified a new species of bacterium, Ideonella sakaiensis, that can “feed” on PET by using it as a source of carbon and energy. The bacterium remains the only known organism with this ability. It literally grows on PET.
“Besides identifying I. sakaiensis, the Japanese scientists discovered that it produces two enzymes and secretes them into the environment,” Silveira explained. “One of the enzymes secreted is precisely PETase. Because it has a certain degree of crystallinity, PET is a polymer that’s very hard to break down. We use the technical term ‘recalcitrance’ to refer to the property certain tightly packed polymers have of resisting degradation. PET is one of them. But PETase attacks it and breaks it down into small units of mono(2-hydroxyethyl) terephthalic acid, or MHET. The units of MHET are then converted into terephthalic acid and absorbed and metabolized by the bacterium.”
All known living beings use biomolecules to survive – all except I. sakaiensis, which is able to use a synthetic molecule manufactured by humans. This means the bacterium is the result of a very recent evolutionary process that has unfolded over the last few decades. The bacterium has adapted to a polymer that was developed in the early 1940s and only began to be used on an industrial scale in the 1970s. PETase is the key to understanding how.
“PETase does the hardest part, which is breaking down the crystal structure and depolymerizing PET into MHET,” Silveira said. “The work done by the second enzyme, which converts the MHET into terephthalic acid, is much simpler, because its substrate consists of monomers that the enzyme can access easily because they’re dispersed in the reaction medium. For this reason, research has focused on PETase.”
The next step was to study PETase in detail, and this is the contribution made by the new research project. “We focused on finding out what gives PETase the capacity to do something other enzymes can’t do very efficiently. We began by characterizing the 3D structure of this protein,” Silveira explained.
“Obtaining the 3D structure means discovering the x, y and z coordinates of each of the thousands of atoms that comprise the macromolecule. Our British colleagues did this using a well-known and widely used technique called X-ray diffraction available at a laboratory very similar to Sirius, now under construction in Campinas.”
Modified enzyme binds better with polymer
Once they had obtained the 3D structure, the researchers began comparing PETase with related proteins. The closest relative is a cutinase of the bacterium Thermobifida fusca that degrades cutin, a sort of natural varnish found on the leaves of plants. Certain pathogenic microorganisms use cutinase to break down the cutin barrier and appropriate nutrients in leaves.
“We found some specific differences in PETase compared with cutinase in the region of the enzyme where the chemical reactions occur, known as the active site. PETase has a more open active site, for example,” Silveira said. “We studied the enzyme’s molecular movements through computer simulations, the part to which I contributed most. While crystal structure, obtained by X-ray diffraction, provided static information, simulations gave us dynamic information and enable us to discover the specific role of each amino acid in the PET degradation process.”
The physics of the molecule’s movements results from electrostatic attraction and repulsion of a great many atoms and from temperature. Computer simulations enabled the researchers to understand more completely how PETase binds and interacts with PET.
“We discovered that PETase and cutinase have two different amino acids at the active site. We then used molecular biology procedures to produce mutations in PETase with the aim of converting it into cutinase,” Silveira said.
“If we could do that, we’d show why PETase is PETase. In other words, we’d find out which components gave it this unique property of degrading PET. However, to our surprise, when we tried to suppress this particular activity of PETase – by trying to convert PETase into cutinase – we produced an even more active PETase. We tried to reduce its activity, and instead we boosted it.”
More computer simulations were required to understand why the mutant PETase was better than the original PETase. Modeling and simulations clearly showed that the alterations produced in the original PETase facilitated the enzyme’s binding to the substrate.
This binding depends both on geometry, with two molecules fitting together like key and keyhole, and on the thermodynamic factors involved in the interactions among the various components of the enzyme and the polymer. The elegant way to describe this is that the modified PETase has “greater affinity” for the substrate.
In terms of a future practical application, of obtaining an ingredient that can digest tons of plastic waste, the study was a great success, but why PETase is PETase remains a mystery.
“Cutinase has amino acids a and b. PETase has amino acids x and y. We thought we could convert PETase into cutinase by replacing x and y with a and b. Instead, we produced an enhanced PETase. In other words, the two amino acids can’t explain the different behavior of these two enzymes. The explanation lies elsewhere,” Silveira said.
Ongoing evolution
Cutinase is an ancient enzyme, while PETase is a modern enzyme resulting from the evolutionary pressure that enabled I. sakaiensis to adapt to an environment containing only or mainly PET as a source of carbon and energy.
A mutation in one of the many bacteria that cannot use this polymer led to the emergence of a species that can. That species reproduced and proliferated much more than the others because it had an abundant food supply. At least, this is the explanation according to standard evolutionary theory.
“The fact that we got a better enzyme by making a small change strongly suggests this evolution is not yet complete,” Silveira said. “There are still new evolutionary possibilities to understand and explore, with the aim of obtaining even more efficient enzymes. The enhanced PETase isn’t the end of the road. It’s just the beginning.”
With a view to application, the next step is to transition from the laboratory scale to the industrial scale, which will require more research related to reactor engineering, process optimization and cost reduction.
The article “Characterization and engineering of a plastic-degrading aromatic polyesterase” (doi: https://doi.org/10.1073/pnas.1718804115) by Harry P. Austin, Rodrigo L. Silveira, Munir S. Skaf et al. can be accessed at: pnas.org/content/early/2018/04/16/1718804115.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





