

A method described in Nature Communications offers improved insight into reactions of enzymes that play key roles in biological processes such as photosynthesis and cell respiration (image: release)
A method described in Nature Communications offers improved insight into reactions of enzymes that play key roles in biological processes.
A method described in Nature Communications offers improved insight into reactions of enzymes that play key roles in biological processes.
A method described in Nature Communications offers improved insight into reactions of enzymes that play key roles in biological processes such as photosynthesis and cell respiration (image: release)
By Karina Toledo
Agência FAPESP – In an article published recently in the journal Nature Communications, researchers based in Brazil and Canada describe a new method for studying the mechanics of chemical reactions catalyzed by metalloproteins, enzymes that contain metal ions bound to the polypeptide chain.
The work is supported by FAPESP through the project “Development and application of computer simulation and spectroscopical analysis to study metalloenzymes and flexible proteins”. The principal investigator is Guilherme Menegon Arantes, co-author of the article and a professor at the University of São Paulo’s Chemistry Institute (IQ-USP) in Brazil.
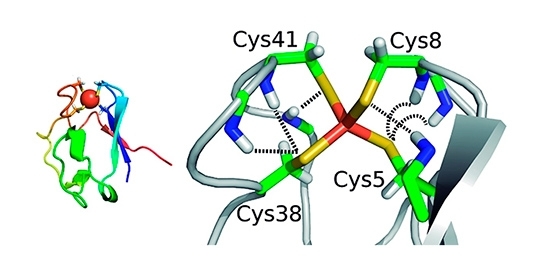
“Metalloproteins, especially those that contain iron and sulfur atoms, are involved in various key biological processes, such as photosynthesis and cell respiration,” Arantes told Agência FAPESP. “Understanding how these enzymes are formed and how their chemical bonds are broken will give us better insight into the biological processes in which they’re involved.”
Because metal atoms have a more flexible electron structure, they are more reactive than purely organic molecules, which consist only of amino acids, he explained. For this reason, metalloproteins are capable of catalyzing more difficult chemical reactions that would not occur in their absence.
According to Arantes, the main challenge for scientists interested in studying metalloproteins has been accessing them in the laboratory. As proteins assume their functional shape, the polypeptide chains fold like origami and, at the same time, roll up in a process known as winding. Because metalloproteins are highly reactive, evolution is believed to have led them to become hidden inside the three-dimensional structure in order to avoid unwanted reactions.
“This facilitates the activity of these enzymes but makes life difficult for researchers who want to study them,” Arantes said. “Until now the methodology consisted basically of dismantling their 3D structure using drastic physical techniques or chemicals. In our study we used a novel methodology that enabled us to unravel the protein only partially, without damaging the polypeptide chain, so that the enzymes could be studied in a situation that more closely resembles those found in living organisms.”
The group, which includes researchers from the University of British Columbia in Canada as well as IQ-USP, is particularly interested in proteins with iron-sulfur centers. Present in all living organisms, these proteins perform a variety of functions, including transferring electrons between other proteins, catalyzing enzyme activity, and regulating genes. The model described in the article used a protein called rubredoxin, which transports electrons in microorganisms.
The Canadian researchers used atomic force microscopy (AFM) to manipulate the protein’s polypeptide chain, stretching it and, at the same time, measuring its movement and the force required for unfolding to occur.
In parallel, the Brazilian group coordinated by Arantes performed a computer simulation of the chemical reaction. “Although the experiment can measure the force and movement required for unfolding and unwinding with a fair degree of precision, it doesn’t offer a microscopic view of the process. The simulation produced a sort of highly detailed film showing the protein’s structure and how chemical bonds are formed and broken. This enabled us to compare and validate the simulation with real data obtained in the experiment,” Arantes said.
The group at IQ-USP performed the simulation using a technique known as hybrid potentials, for which the 2013 Nobel Prize in Chemistry was awarded to Martin Karplus (Harvard University), Michael Levitt (Stanford University) and Arieh Warshel (University of Southern California).
“I’ve been working with this technique since my PhD with FAPESP’s support,” Arantes said. “It uses a combination of molecular and quantum mechanics to describe how chemical bonds are formed or broken in proteins and other complex molecular systems.”
Although quantum mechanics is the most appropriate theory to describe this kind of phenomenon, he explained, its use becomes unviable when the system to be studied is as complex as a protein made up of thousands of atoms.
“Even a supercomputer can’t perform such huge calculations, so we use approximations,” he said. “Quantum mechanics is used only to study the catalytic site of the enzyme. The rest of the molecule, which isn’t involved in the chemical reaction, is described using molecular mechanics, without detailing the electron structure.”
A previous paper published in the journal Angewandte Chemie described how the group simulated the dissociation of iron-sulfur clusters in rubredoxin during unfolding, without the presence of any other chemical reagent.
In the study described by the more recent paper, the researchers added a nucleophilic agent, which bonds to the iron atom, and an electrophilic agent, which bonds to the sulfur atom.
“The experiment measured the change in force required to break the natural bond between iron and sulfur in the presence of other competing agents,” Arantes said. “We designed the simulation to understand how the chemical reaction works and how it’s facilitated by partial unfolding of the protein and by the presence of chemical agents.”
This hybrid technique, he added, is a powerful tool for studying the reactions and stability of several metalloproteins. “If we can understand the reactivity of these metal centers more profoundly, we’ll be able to try to find ways to control electron leaks, which are responsible for some 90% of the free radicals produced inside cells,” he said. “Free radicals react with biomolecules and may impair cell functioning.”
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





