

Schematic illustration providing a graphic summary of the study (credit: Archives of Biochemistry and Biophysics)
A yeast experiment conducted at a FAPESP-supported research center shows how deficiencies in the proteasome, which is responsible for eliminating damaged and non-functional proteins, affect the metabolism of mitochondria, the organelles that generate energy for cells.
A yeast experiment conducted at a FAPESP-supported research center shows how deficiencies in the proteasome, which is responsible for eliminating damaged and non-functional proteins, affect the metabolism of mitochondria, the organelles that generate energy for cells.
Schematic illustration providing a graphic summary of the study (credit: Archives of Biochemistry and Biophysics)
Agência FAPESP* – A study by the Center for Redox Processes in Biomedicine (Redoxoma) led by Marilene Demasi from the Butantan Institute (São Paulo, Brazil) presents a valuable new experimental model for investigating the interaction between the proteasome and mitochondrial function. In eukaryotic cells, the proteasome is a protein complex responsible for eliminating damaged and nonfunctional proteins, thereby helping to maintain cellular balance and proper functioning.
In recent years, studies have revealed that the proteasome and mitochondria are more closely connected than previously thought. The proteasome plays a role in the quality control of proteins destined for the mitochondria, while mitochondrial metabolism affects the efficiency with which proteins marked for destruction are degraded.
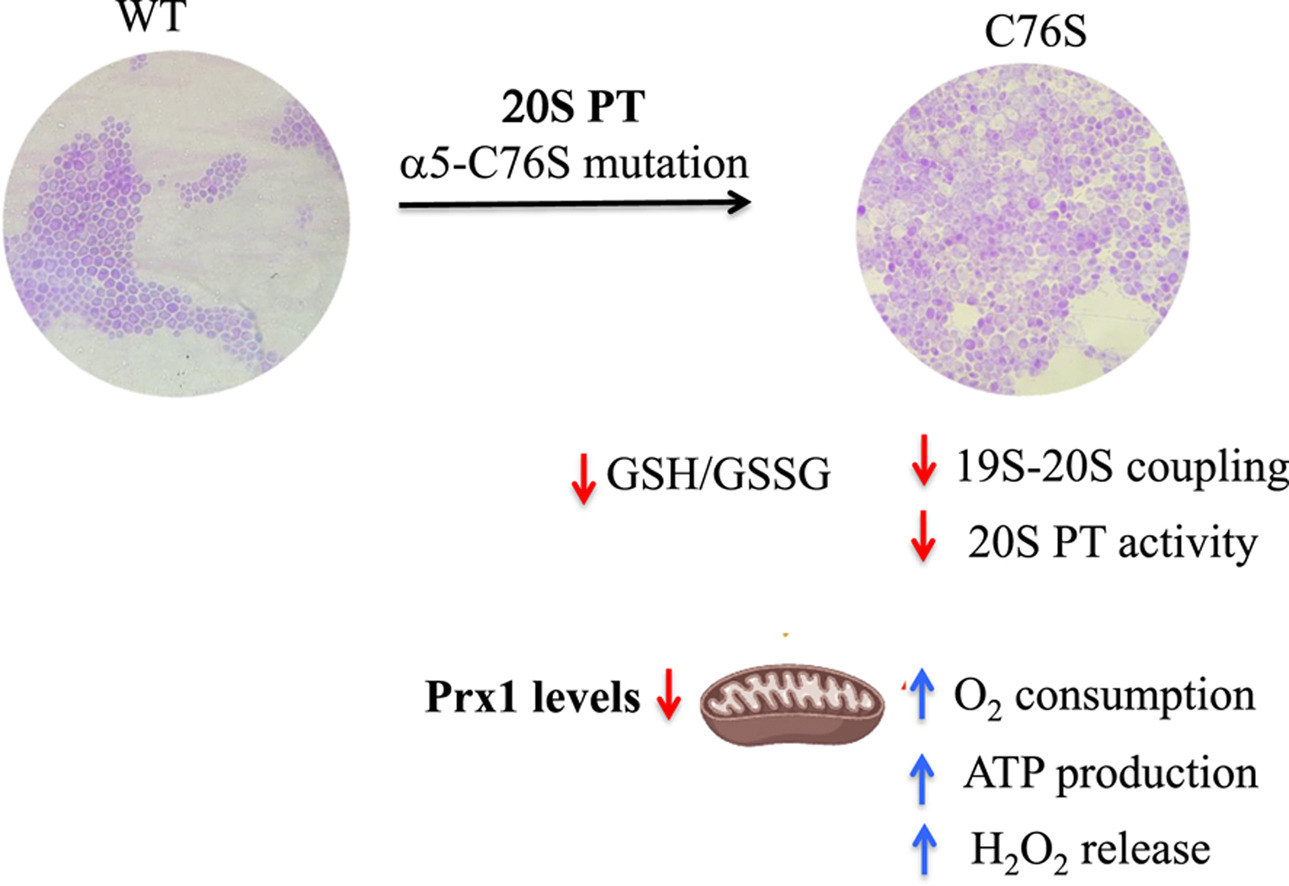
Redoxoma, a Research, Innovation and Dissemination Center (RIDC) of FAPESP based at the University of São Paulo’s Institute of Chemistry (IQ-USP) conducted research focusing on the effects of proteasome dysfunction in the C76S mutant strain of the yeast Saccharomyces cerevisiae. The study revealed that deficiency in this system leads to increased mitochondrial oxidative stress. This was evidenced by increased hydrogen peroxide (H2O2) release and a lower peroxiredoxin 1 (Prx1) concentration. Prx1 is a crucial enzyme in the removal of peroxides. In mammals, mitochondrial Prx3 is equivalent to Prx1 in yeast.
“The most important thing about this work is that we’ve a yeast strain that can serve as a model for investigating proteasome deficiency in relation to mitochondrial metabolism, a model that didn’t yet exist in the literature,” Demasi points out.
The study was published in an article in the journal Archives of Biochemistry and Biophysics.
Next steps
The researchers are now working to understand why Prx1 levels decrease in cells with compromised proteasomes. “We don’t yet know if there was a decrease in Prx1 gene expression, which is possible, since the proteasome also plays a role in gene transcription regulation, or if the protein oxidizes more. It may hyperoxidize and, as a result, be degraded more. Perhaps the excess peroxide is promoting its continuous degradation,” says the researcher at the Butantan Institute.
To answer these questions, the group plans to conduct comparative transcriptome and proteomic analyses of the wild and mutant strains cultivated under respiratory conditions. The goal is to establish this strain as a model for studying the role of the ubiquitin-proteasome system in cell metabolism.
The article “Decreased levels of Prx1 are associated with proteasome impairment and mitochondrial dysfunction in the yeast Saccharomyces cerevisiae” can be read at: www.sciencedirect.com/science/article/abs/pii/S0003986125001195?via=ihub.
* With information from Maria Celia Wider, from Redoxoma.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





