

3D structure of ribosomal complex. Proteins modified by K63 ubiquitin chains identified using quantitative mass spectrometry are shaded in blue (image: Silva et al., 2015/Nature Structural & Molecular Biology)
Paper published in Nature Structural & Molecular Biology describes discovery that could pave the way to new treatment for cancer and neurodegenerative diseases such as Parkinson's and Alzheimer's.
Paper published in Nature Structural & Molecular Biology describes discovery that could pave the way to new treatment for cancer and neurodegenerative diseases such as Parkinson's and Alzheimer's.
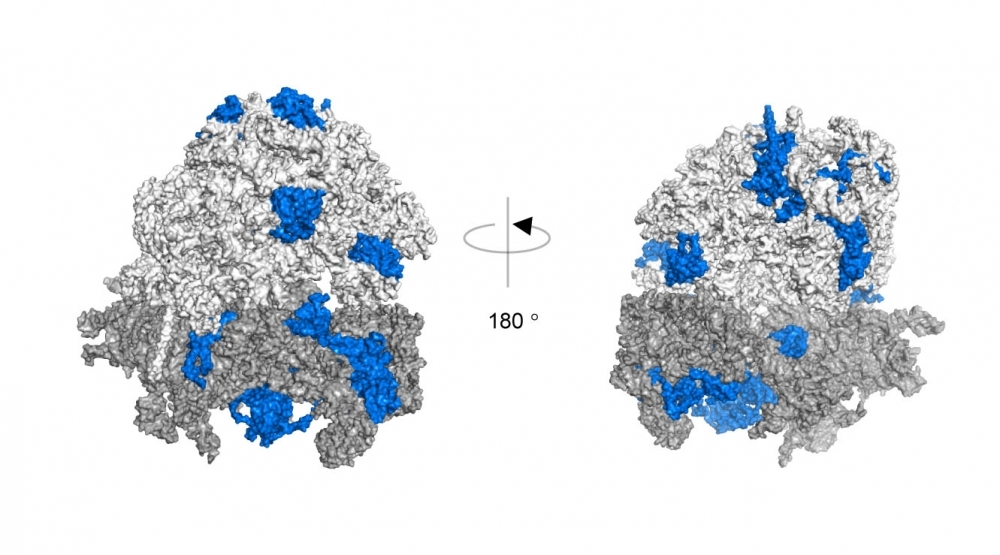
3D structure of ribosomal complex. Proteins modified by K63 ubiquitin chains identified using quantitative mass spectrometry are shaded in blue (image: Silva et al., 2015/Nature Structural & Molecular Biology)
By Karina Toledo
Agência FAPESP – In a paper published by the journal Nature Structural & Molecular Biology, researchers at New York University (NYU) and Harvard University in the United States describe a new insight into the mechanisms used by cells to defend themselves from oxidative stress.
This biological condition is characterized by an increase in the levels of free radicals and other oxygen-reactive species in the cellular environment during various physiological processes, such as inflammation and aging. This condition may also result from exposure to pollution, tobacco smoke, radiation and chemicals in industrialized foods and beverages.
In excess, oxidants damage nucleic acids, proteins and other molecules important to cellular function. If this process becomes chronic, it can lead to the development of tumors and neurodegenerative diseases such as Parkinson’s and Alzheimer’s.
“We describe a new cellular signaling pathway in response to oxidative stress that has been completely unknown until now. The discovery extends our understanding of how cells respond to this kind of aggression and may reveal new therapeutic targets for exploration in the treatment of several diseases,” said Gustavo Monteiro Silva, a post-doctoral fellow at NYU and first author of the paper. Daniel Finley of Harvard University and Christine Vogel, also of NYU, are co-authors.
Silva began his research in the field when he was preparing his PhD thesis at the University of São Paulo’s Bioscience Institute (IB-USP) in Brazil, with support from FAPESP.
Supervised by Luis Eduardo Soares Netto and Marilene Demasi, he investigated the mechanism whereby an intracellular protein complex called the proteasome is regulated to degrade proteins damaged during oxidative stress (read more at http://agencia.fapesp.br/15583).
After completing his doctoral research, Silva partnered with Vogel in proteomics and systems biology research to achieve a better understanding of the cellular response to stress. The researchers observed that as well as regulating the removal of damaged proteins by the proteasome, cells can defend themselves by controlling the production of new cells that play a key role in stress survival.
Both defense mechanisms are associated with a signaling pathway mediated by a protein called ubiquitin. This molecule forms chains (polyubiquitin) that bond to target proteins in a process known as ubiquitination.
Not long ago, it was believed that ubiquitination served only to signal to the proteasome that a given protein should be degraded, and for this reason, it was nicknamed the “kiss of death.”
More recent studies, however, have shown that polyubiquitin chains can perform other functions, depending on how the ubiquitin molecules interconnect and organize themselves spatially.
“Protein ubiquitination in response to oxidative stress is essential for cells, but scientists have debated its role for over 30 years,” Silva said. “Knowing that there were different ubiquitination chains, I decided to investigate the types, targets and importance of these chains for cells’ responses to oxidative stress.” Trials were performed using Saccharomyces cerevisiae yeast cells. To induce oxidative stress, the researchers initially treated the cells with hydrogen peroxide.
With the aid of mass spectrometry techniques and antibodies specific to the various chain types, the scientists observed the expected increase in K48 polyubiquitin chains (linked by the amino acid residue lysine 48 in ubiquitin), which is associated with protein degradation.
The research also produced an entirely novel finding: a rapid increase in an alternative polyubiquitin chain type known as K63. This proved to be a specific response to the oxidative stress induced by peroxides. When these oxidizing substances were removed from the culture medium, the number of K63-type chains fell rapidly in a highly regulated process.
“We then decided to investigate this alternative chain, K63, in greater depth, and using quantitative proteomics, we found that it modifies certain ribosomal proteins, making this structure more stable and favoring the synthesis of proteins important to the antioxidant response,” Silva said.
Therapeutic targets
For the ubiquitination process to take place, there has to be a cascade of reactions catalyzed by various enzymes. In Silva’s view, identification of the enzymes specifically involved in this pathway enables potential targets to be found for the development of drugs against disorders and diseases associated with oxidative damage.
“We observed that peroxides reversibly inhibit the action of a deubiquitinating enzyme called Ubp2 [whose function is to remove ubiquitin molecules from its targets], thereby favoring accumulation of the K63 chain,” Silva said.
In experiments using a mutant yeast strain incapable of forming K63 ubiquitin chains, the researchers observed increased oxidative damage and reduced protein production, and the mutant cells became more sensitive to oxidative stress.
“If we can achieve a better understanding of the function of these enzymes and how this chain regulates protein production, we can try to modulate the cellular response to oxidative stress, both to promote cell death when treating tumors and to make cells more resistant, which would be interesting for the treatment of neurodegenerative diseases,” Silva said.
The group also performed trials with mouse neuronal cells, again observing that K63 polyubiquitin chains were formed in response to stress. According to Silva, the next step is to find out more about how this defense mechanism operates in mouse cells, based on the knowledge obtained from the studies using yeast. In the future, the group plans to perform trials with human cells.
The paper “K63 polyubiquitination is a new modulator of the oxidative stress response” (doi:10.1038/nsmb.2955), by Gustavo M. Silva et al, was published in Nature Structural & Molecular Biology and can be read by subscribers at www.nature.com/nsmb/journal/vaop/ncurrent/full/nsmb.2955.html.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





