
Understanding the natural interaction of two proteins to inhibit bacteria cell division – and in this manner, possibly preventing bacterial proliferation - is the topic of a proposal approved in the RFP launched by the FAPESP–SMOLBnet 2.0
Understanding the natural interaction of two proteins to inhibit bacteria cell division – and in this manner, possibly preventing bacterial proliferation - is the topic of a proposal approved in the RFP launched by the FAPESP–SMOLBnet 2.0
Understanding the natural interaction of two proteins to inhibit bacteria cell division – and in this manner, possibly preventing bacterial proliferation - is the topic of a proposal approved in the RFP launched by the FAPESP–SMOLBnet 2.0

Understanding the natural interaction of two proteins to inhibit bacteria cell division – and in this manner, possibly preventing bacterial proliferation - is the topic of a proposal approved in the RFP launched by the FAPESP–SMOLBnet 2.0
By Mônica Pileggi
Agência FAPESP – Understanding the natural interaction of two proteins to inhibit bacteria cell division – and in this manner, possibly preventing bacterial proliferation - is the topic of a proposal approved in the RFP launched by the Network for Structural Molecular Biology–SMOLBnet 2.0.
Coordinated by Professor Frederico José Gueiros Filho of the Biochemistry Department at the Universidade de São Paulo’s (USP) Chemistry Institute, the SMOLBnet2.0 project “Combining genetics and NMR to dissect fundamental protein-protein interactions for complex bacterial division” intends to study how a protein known as FtsZ interacts with two others found in the bacteria: MinC and MciZ.
Gueiros Filho’s laboratory team has been uncovering the inner workings of bacteria cells for years, specifically the role of proteins that promote cell division.
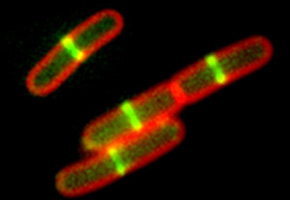
“There is a group of roughly 20 proteins specialized in executing division of bacteria cells. The maestro of them all is FtsZ. It is interesting because it has the ability to pinpoint the exact place that the cell needs to divide. By forming a ‘band’ on the internal part of the bacteria, it attracts other division promoting proteins to the ring it has just formed,” Gueiros explains to Agência FAPESP.
It is the proteins attracted by FtsZ that alter the external wall of the bacteria. At the moment of division, the bacteria’s wall stops expanding at its height and grows inward, forming a disco or septum, which separates bacteria into two daughter cells.
One important aspect of the division process is that it must be well regulated and cannot occur at just any moment. Because of this, the bacteria itself houses proteins that function as natural division inhibitors. One example is the MinC protein, which connects to the FTsZ protein and impedes the latter from forming the division ring at the wrong time or place.
In studying the effects of the MinC protein on FtsZ, the team first hopes to understand the natural process of cell division inhibition. “There is no doubt, therefore, that the information obtained can serve as inspiration for development of an antibiotic capable of impeding the proliferation of bacteria. It is as if it were a reverse engineering process: we study the bacteria inhibitor and utilize the principles learned to make our own inhibitor,” explains Gueiros.
The initial tests of the project, which are slated to begin in March, will be conducted with Bacillus subtilis. “This bacterium, which lives in the ground and is not pathogenic, is what we call a model system. It is an easily studiedorganism which serves as an example for others. As the division mechanism is similar in all bacteria – all have FtsZ -, what we have learned with Bacillus subtilis is applicable to many other species, including pathogens,” he says.
Complementary forms
Proteins are formed by diverse molecules of amino acid chains. Their three-dimensional globular format is reminiscent of a spherical coral, with cavities and saliencies on its surface. In order to connect to each other, these proteins must have not only a chemical affinity, but complementary shapes.
In order to discover which parts of FtsZ and MinC are involved in this fitting, the team at USP’s Chemistry Institute has identified more than a dozen mutant strains of FtsZ, or rather variations in which an amino acid of this protein was substituted and as a result are no longer inhibited by MinC.
“When we found this variation, we learned about how the proteinfunctions. We observed that the mutations are very similar to each other. This gives us an indication that the region is important for one protein to connect to another,” affirms Gueiros.
According to him, the next step will be to refine the study using nuclear magnetic resonance (NMR), a high precision technique through which one can observe which atoms are involved in the interaction of the proteins studied.
“Nuclear magnetic resonance allows one to confirm what role each one of these mutants represents at the site of interaction between FtsZ and MinC. The technique will also expand what has already been discovered without the use of this technology. We believe that there are other amino acids involved in this interaction,” he notes.
The study involves collaboration with researcher Ana Carolina de Matos Zeri, coordinator of the Multiuser Nuclear Magnetic Resonance Laboratory at the National Bioscience Laboratory (LNBio), which is part of the National Energy and Matter Research Center (CNPEM).
The group also intends to use resonance to determine the three-dimensional structure of MciZ, one of the three proteins for which atom placement and inhibition mechanisms are unknown.
In addition to coordinators, the project will involve doctoral students and one post-doctoral researcher. There is interest in incorporating another post-doctoral candidate.
“There will be a synergy of expertise, like bacterial cell division and nuclear magnetic resonance. We intend to unite these two specialties to make better scientific observations than we could alone,” affirms Gueiros. Interested candidates can submit curriculum vitae to fgueiros@iq.usp.br.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





