

Researchers at São Paulo State University have completed the first mtDNA sequencing of one of Brazil’s more than 120 species of carnivorous plants (photo: utricle from Utricularia reniformis A.St.-Hil. with captured arthropod larva / release)
Researchers at São Paulo State University have completed the first mtDNA sequencing of one of Brazil’s more than 120 species of carnivorous plants.
Researchers at São Paulo State University have completed the first mtDNA sequencing of one of Brazil’s more than 120 species of carnivorous plants.
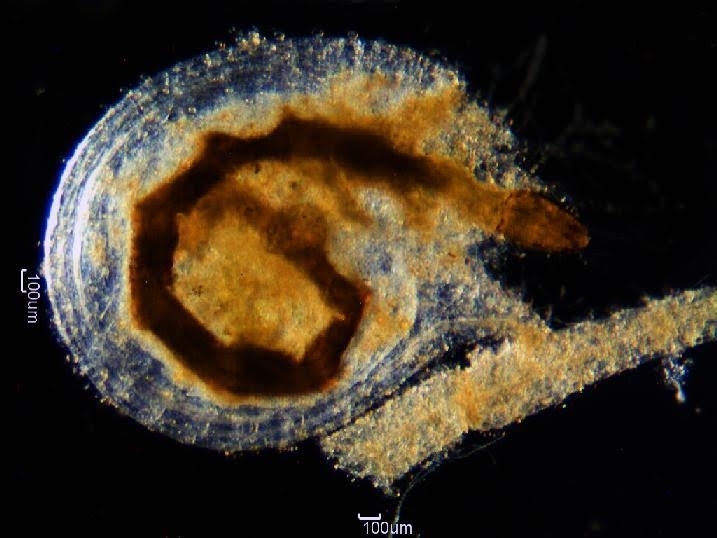
Researchers at São Paulo State University have completed the first mtDNA sequencing of one of Brazil’s more than 120 species of carnivorous plants (photo: utricle from Utricularia reniformis A.St.-Hil. with captured arthropod larva / release)
By Peter Moon | Agência FAPESP – Some 130 species of carnivorous plants are found in practically all biomes in Brazil. To understand how the evolutionary dynamics of this fascinating group of plants unfolded, a necessary condition is the study of their DNA, but none of these Brazilian species has had its complete genome sequenced.
This is now changing thanks to the work of a group of researchers at São Paulo State University (UNESP) who are sequencing the whole genome (nuclear, chloroplast and mitochondrial DNA) of a carnivorous plant, Utricularia reniformis, that is endemic to Brazil.
The first results, published in 2016, encompassed the plant’s plastome, or its chloroplast genome (cpDNA). A second study, focusing on the mitochondrial genome (mtDNA), has just been published, also in PLOS ONE. The next step is sequencing of the nuclear genome (nDNA), currently in progress.
The research is led by bioinformaticist Alessandro de Mello Varani and biologist Vitor Fernandes Oliveira Miranda, both affiliated with the School of Agrarian & Veterinary Sciences, UNESP Jaboticabal, and is supported by FAPESP.
The 130-odd species of carnivorous plants identified to date in Brazil do not descend from a common ancestor. Carnivory in plants evolved on more or less nine different occasions in five different orders of angiosperm.
The order with the most carnivorous plants is Lamiales, which is also includes lavender, lilac and snapdragon as well as plants of culinary importance such as olive, jasmine, mint, rosemary, sunflower, sage and basil.
In the order Lamiales, the family with the most carnivorous species (370) is Lentibulariaceae, and some 230 of these species belong to the genus Utricularia (bladderworts), which comprises aquatic and terrestrial plants. Utricularia reniformis is a terrestrial bladderwort that grows in moist Atlantic Rainforest areas.
The method used by U. reniformis to capture its prey involves a tiny baglike bladder called a utricle. Inside the utricle is a fluid that serves to digest microcrustaceans trapped by the plant. Once caught, the prey cannot escape; it dies and is digested and absorbed by the carnivorous plant.
All this takes place on such a small scale that it is invisible to the naked eye. “The utricles are tiny and can only be analyzed in detail under a microscope,” Varani said.
“The smallest plant genomes are believed to be those of some species of Lentibulariaceae. The nuclear genomes (nDNA) of animals are better conserved, meaning they vary little among species of the same genus or family compared with plant genomes.”
The nuclear genomes of humans and chimpanzees, for example, differ by only 1.5%. Plants of the same species but from different populations, however, may have strikingly different nuclear genomes.
“They vary immensely within the same genus and even across different individuals in the same species,” Varani said. “In Lentibulariaceae, for example, the size of the nuclear genome can vary from 61 million to 1.6 billion bases (nucleotides) within the same family. There are species with genomes that are 25 times smaller than those of others. So Lentibulariaceae are excellent candidates for studies of genomic contraction and expansion.”
Besides the varying sizes of the genomes of plants in the same genus or species, the order of the genes in their DNA spirals also changes. It is as if the millions of nucleotides in a plant’s nuclear DNA had been poured into a blender and mixed up into a new sequence, without impairing the functionality of the resulting DNA or its belonging to the same species.
“Plant genomes fluctuate in size and in terms of the order of the bases in chromosomes,” Varani said. “Working with the nuclear genomes of plants is therefore a challenge from the standpoint of bioinformatics.”
It is a complicated task, yet a feasible one, thanks to the advances achieved in sequencing techniques, and especially in bioinformatics, the computational tool used to classify and compare the dozens of bases in each genome. The challenge is determining which portions of the genome correspond to functional genes.
“You have to determine where the sequence of bases for a given gene begins and where it ends. And you have to discover which of the different DNA sequences taken from members of the same species are genes inherent in that species,” Varani said. “In this specific case, the goal is to discover which genes are the ones that determine the carnivorousness of the plant U. reniformis.”
Chloroplast DNA
The main way in which plants differ genetically from other types of organisms is that in addition to nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) – which are found in animals as well as plants – they also have a third genetic reservoir not found in animals: the chloroplast genome (cpDNA), or plastome. Chloroplasts are the organelles responsible for photosynthesis.
Symbiotic theory says that organelles like mitochondria and chloroplasts are inherited from ancient microorganisms that invaded or were absorbed by larger bacteria billions of years ago in the Proterozoic. Instead of destroying each other, they established a symbiotic relationship in which the bacterial ancestors of mitochondria and chloroplasts took on the key role of energy generation in the host’s metabolism: energy from food in the case of mitochondria, energy from solar radiation in that of chloroplasts.
Because mitochondria and chloroplasts were independent microscopic beings in the remote past, these organelles have conserved interior DNA sequences that descended directly from the DNA of those tiny Proterozoic invaders.
Whole-genome sequencing of plants entails the sequencing of nuclear DNA (nDNA), with tens of millions of bases, and mitochondrial DNA (mtDNA) and chloroplast DNA (cpDNA), with only a few thousand bases each.
“Sequencing the chloroplast DNA was easiest. We discovered many interesting points, such as the fact that in U. reniformis, many of the chloroplast genes that regulate photosynthesis were deleted. They’d disappeared from the cpDNA, an interesting discovery since these same genes are intact in aquatic species,” Varani said.
“Carnivorous plants perform photosynthesis. However, carnivory is also a way of obtaining nutrients. The loss of those genes may have to do with adaptation to carnivory and to a terrestrial habitat. We just don’t know at this time.”
A key discovery that the researchers made in mtDNA sequencing was that some of the genes deleted from the cpDNA had been transferred to the mtDNA but appeared not to be functional there.
“We’re now studying gene transfer between these organelles and trying to understand this process from an evolutionary standpoint,” Varani said. “We compared the cpDNA and mtDNA of U. reniformis with the genomes of these organelles in other terrestrial species in the same genus and found the same tendency to lose genes from the cpDNA, which had been transferred to the mtDNA in all cases. What’s interesting is that this tendency appears not to occur in aquatic species, suggesting that Lentibulariaceae can also be used as a model for studying how plants adapted to aquatic habitats.”
The article “The mitochondrial genome of the terrestrial carnivorous plant Utricularia reniformis (Lentibulariaceae): Structure, comparative analysis and evolutionary landmarks,” by Saura R. Silva, Danillo O. Alvarenga, Yani Aranguren, Helen A. Penha, Camila C. Fernandes, Daniel G. Pinheiro, Marcos T. Oliveira, Todd P. Michael, Vitor F. O. Miranda and Alessandro M. Varani, can be read at doi.org/10.1371/journal.pone.0180484.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





