
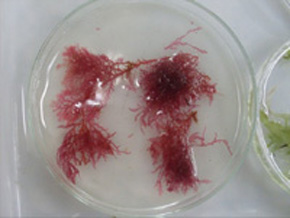
With the help of molecular tools such as DNA barcoding, researchers have identified nearly ten new species (Wikimedia)
With the help of molecular tools such as DNA barcoding, researchers have identified nearly ten new species.
With the help of molecular tools such as DNA barcoding, researchers have identified nearly ten new species.

With the help of molecular tools such as DNA barcoding, researchers have identified nearly ten new species (Wikimedia)
By Karina Toledo, in London
Agência FAPESP - With the help of molecular techniques such as gene sequencing and DNA barcoding, scientists at the University of São Paulo (USP) are completing the most complete survey conducted to date regarding the biodiversity of red macroalgae in the state of São Paulo.
The study is being conducted under the scope of a Thematic Project led by Mariana Cabral de Oliveira of the USP Biosciences Institute.
Some of the findings were presented September 26 during FAPESP Week London. The event was held by FAPESP with support from the British Council and the Royal Society.
“Multicellular algae, or macroalgae, are divided into three groups: red, brown and green. We decided to study the red algae because it comprises the most diverse group found along the Brazilian coast and also has more economic significance,” explained Oliveira.
In addition to serving as food, mainly in Asian cuisine, most of the red macroalgae produce gelatins – such as agar and carrageenan – that are used as frequently in the food, cosmetics and pharmaceutical industries as they are in the biotechnology sector. These algae also produce other by-products of commercial interest, such as dyes and molecules used in agriculture to increase plant growth.
“Algae, in general, are also very important to the ecosystem because in addition to acting as a base of the marine food chain, they produce nearly half of all oxygen on the planet,” the researcher said.
Finally, Oliveira emphasized the importance of studying algae for understanding of the evolution of life on Earth. “They form very diverse lines, and there are groups that have evolved independently. To identify an ancestor that gave rise to all algae, we would have to go back nearly two billion years. Yet, animals and fungi separated one billion years ago. In other words, from a phylogenetic standpoint, we humans are closer to fungi than certain groups of algae are to each other,” she commented.
The collection
To conduct the survey, the researchers collected samples from the coastal regions of the state of São Paulo as well as on the islands. This work was accomplished with help from divers and the collaboration of Mutue Fujii of the Botanical Institute. Specimens of fresh water from São Paulo rivers were collected in an offshoot of the thematic project, led by Professor Orlando Necchi Jr. of the Universidade Estadual Paulista (Unesp) of São José do Rio Preto.
“The red marine macroalgae are limited to the coastal region because they grow attached to the hard substrate that extends to a depth of nearly 20 meters in the state of São Paulo,” said Oliveira.
The DNA of all the organisms collected was extracted for analysis and then deposited in the herbaria of the Department of Botany at the USP/Biosciences Institute, the Botanical Institute, and the Institute of Biosciences, Letters and Exact Sciences at Unesp of São José do Rio Preto, where it is available for other groups who may be interested in studying it. The genetic markers sequenced during the species identification project are also stored in a publicly accessible database that can be accessed at www.boldsystems.org.
“We are realizing that traditional taxonomic techniques based on the morphology of the species alone are not enough to achieve a proper classification in some situations. There are cases in which we found two organisms with varying morphology and, after examining them more thoroughly, we saw that they in fact belonged to the same species but had greater phenotypic plasticity,” said Oliveira.
Often, the opposite occurs, added the researcher. Two specimens that seem to belong to the same species end up being different upon molecular analysis.
To conduct these analyses, the researchers made use of whole gene sequencing through traditional techniques as well as DNA barcoding, which is a method that analyzes only a small section of the gene. The latter is thus faster and cheaper and allows the analysis of a larger number of samples.
“First, we used DNA barcoding to define the taxonomic groups (i.e., separate the species) and then we selected individuals to perform the molecular marking – in other words, to sequence a whole gene. One of the genes we use to differentiate the species is rbcL, which encodes the large subunit of the enzyme RuBisCO (ribulose-1,5-bisphosphate carboxylase oxygenase),” she explained.
The findings
According to Oliveira, before the study began, 190 species of red macroalgae had already been described in the state of São Paulo. By the time the project is over, the final number could be closer to 240.
“There are over a dozen new species that have never been described before. There are also a series of new occurrences, in other words, species that have already been described in other regions but have not appeared in citations for the coast of São Paulo. Right now, we’re summarizing the results, and the number is still not final,” Oliveira said.
The researcher went on to say that the state of São Paulo is the most well-studied Brazilian state in terms of algal diversity, and work in this area has been conducted since the 1950s.
During her presentation at FAPESP Week London, Oliveira also spoke about the Brazilian Barcode of Life (BrBOL) project, a consortium designed to map Brazilian biodiversity through the use of molecular tools.
“It’s a Brazilian initiative supported by the National Council for Scientific and Technological Development (CNPq). The aim is to participate in an international effort to create a molecular label, or barcode, for every species on the planet. I coordinate the marine group, which includes algae and some invertebrate groups,” she added.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





