

The automated system developed by Brazilian researchers is based on an analysis of patterns of molecules in patient blood plasma (image: Anderson Rocha)
The automated system developed by Brazilian researchers is based on an analysis of patterns of molecules in patient blood plasma.
The automated system developed by Brazilian researchers is based on an analysis of patterns of molecules in patient blood plasma.
The automated system developed by Brazilian researchers is based on an analysis of patterns of molecules in patient blood plasma (image: Anderson Rocha)
By Karina Toledo | Agência FAPESP – Developed in Brazil, a low-cost method of diagnosing COVID-19 in approximately 20 minutes that does not require imported reagents is described in a paper published on the platform medRxiv. The paper is a preprint and has not yet been peer-reviewed.
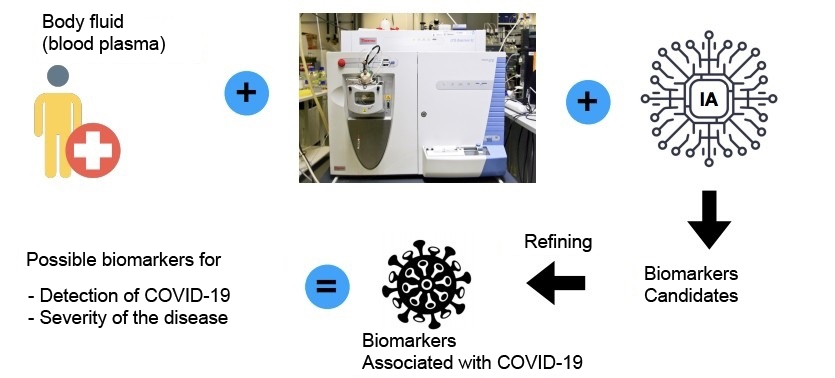
The system uses artificial intelligence algorithms to analyze patient blood plasma samples in search of patterns of molecules characteristic of the disease. According to the authors, it can also analyze confirmed cases of the disease to detect whether a subject risks developing severe manifestations such as respiratory failure.
The project is supported by FAPESP and involves researchers at the University of Campinas (UNICAMP) and the University of São Paulo (USP), as well as collaborators in Manaus, the state capital of Amazonas.
“In tests performed to validate the methodology, we distinguished between positive and negative samples with a success rate in excess of 90%. We also distinguished between severe and mild cases with 82% accuracy. We have applied for certification with ANVISA [the national health surveillance agency],” Rodrigo Ramos Catharino, a professor at UNICAMP and principal investigator for the project, told Agência FAPESP.
When ready, he added, the diagnostic test will cost approximately 40 Brazilian reais per sample, or half the cost of a molecular test by the RT-PCR method, considered the gold standard for the diagnosis of COVID-19.
The project is being conducted at the Innovare Biomarker Laboratory in UNICAMP’s School of Pharmaceutical Sciences as part of the PhD research of Jeany Delafiori. It is also part of a research line combining metabolomics and machine learning in pursuit of markets to assist in the diagnosis of diseases such as Zika, hemorrhagic dengue, cystic fibrosis, diabetes and/or other metabolic disorders.
The researchers work in partnership with the Complex Data Inference Laboratory (RECOD) in UNICAMP’s Institute of Computing (IC), led by Anderson Rocha, with Luiz Claudio Navarro collaborating.
“We recruited 728 patients for the project, among whom 369 had been diagnosed with COVID-19, confirmed clinically and by RT-PCR,” Delafiori said. “Samples from uninfected subjects were used for comparison as a sort of control group. In the case of some patients who developed complications and had to be hospitalized, a second blood sample was taken. Generally, there were individuals with mild and severe symptoms among the patients with confirmed cases.”
All samples were analyzed with the aid of a mass spectrometer, which identifies substances in biological fluids based on their molecular weight. As the researchers explained, the molecules found in blood plasma point to the metabolic processes active in the organism.
“We focused on low molecular weight molecules, such as amino acids, small peptides and lipids. These are the end-products of metabolic processes and are therefore directly linked to the symptoms manifested by patients when the samples were collected,” Delafiori said.
Some samples were then used by the IC-UNICAMP group to train the machine learning algorithm to recognize metabolite patterns found in positive and negative cases and to distinguish between mild and severe cases. Others were used in a blind test designed to evaluate the accuracy of the analysis performed by the system.
According to the paper, the diagnostic specificity and sensitivity were 97.6% and 83.8%, respectively, in the blind test, while the severity risk assessment achieved 76.2% specificity and 87.2% sensitivity.
“Sensitivity refers to the method’s capacity to detect the presence of the virus and specificity to its capacity to distinguish between this and other diseases. When combined, these two parameters determine accuracy,” Delafiori said. “We continue to work to improve the accuracy of the test as our collaborators collect fresh samples from patients.”
According to Rocha, the algorithm incorporates knowledge as it analyzes samples and hence improves its performance over time. “The accuracy is currently approximately 90%, and it will tend to become more accurate when thousands of patients have been analyzed,” Delafiori said.
The IC-UNICAMP group has also written software that automates the entire process of analysis, producing a final report that tells the physician whether the patient has COVID-19 and risks developing complications.
“These biomarkers that predict the progress of the disease can help primary care medical teams decide whether patients who test positive can be kept in isolation at home or should be transferred to a higher level of care, for example,” said Rinaldo Focaccia Siciliano, another coauthor of the paper. Siciliano is an attending physician in the Infectious and Parasitic Disease Division of Hospital das Clínicas (HC), the general and teaching hospital run by the University of São Paulo’s Medical School (FM-USP), and in the Hospital Acquired Infection Control Unit at its Heart Institute (INCOR).
According to Siciliano, the method has proven accurate in detecting both mild cases during the first few days of symptom presentation and more severe cases in which the patient displays respiratory discomfort on admission to the hospital. “The advantage of having several centers participating in the project with different profiles is sample variability, so that the methodology can be applied in different scenarios, such as outpatient and hospital care,” he said.
Another advantage of the method, he added, is that it can diagnose the disease early on by analyzing blood samples, which are easier to collect than the nasal and throat swabs required for RT-PCR. “Nasopharyngeal swab collection requires a well-trained team and a suitable room, given the risk of aerosol transmission of the virus. The currently available blood test can detect antibodies only a few days after symptoms appear,” he said.
Optimized model
Most laboratory blood tests analyze the levels of a few substances, whereas the computational system devised by the IC-UNICAMP group can simultaneously analyze thousands of variables and extract direct and cross connections, such as finding substances with augmented or diminished levels in subjects with a particular disease.
“To make this possible, we have worked for the last three years on the development of a mathematical model that is explainable, meaning it enables us not only to make accurate predictions but also to know which variables the system is looking at to make these predictions. As a result, once we have identified a first set of biomarkers, we can select the most significant among them and optimize the analytical process. In addition, the data generated can be used by metabolomics researchers to investigate the mechanism of the disease,” Navarro said.
In the case of COVID-19, the group identified a set of 30 metabolites that can be considered a signature of the disease. According to Delafiori, a positive diagnosis was associated, for example, with diminished levels of lysophosphatidylcholines – glycerol-derived phospholipids that contain phosphate in their structure. “These molecules are precursors of pulmonary surfactants [compounds that reduce surface tension in the alveoli, preventing lung collapse during expiration] and protect the lungs from opportunistic infections. Diminished levels of these compounds have been reported in patients with severe acute respiratory syndrome,” she said.
Diminished levels of cholesterol derivatives were also observed in positive cases, and this decrease was especially pronounced in patients who progressed to the most severe form of the disease. “Some studies have reported a reduction in cholesterol as COVID-19 patients deteriorate toward an adverse outcome,” Delafiori said.
Glycerolipid levels, previously reported to be dysregulated in severe acute respiratory syndrome, were augmented in samples from COVID-19 patients.
“Biochemical validation of biomarkers has enabled us, for example, to discard molecules associated with the use of anti-inflammatory medication and not with the disease itself. Subsequently, we combined the remaining variables into pairs. The novel technique we’ve introduced into the model increases the accuracy of the analysis and enables it to be performed with different mass spectrometers,” Navarro said.
For Catharino, the methodology can be used by any public or private laboratory equipped with a mass spectrometer. While awaiting ANVISA’s approval, the researchers plan to increase the diversity of the samples analyzed in the context of the research to further improve the system’s performance.
Researchers at Amazonas State University (UEA), Dr Heitor Vieira Dourado Tropical Medicine Foundation (FMT-HVD), Amazonas Health Surveillance Foundation (FVS-AM), Leônidas & Maria Deane Institute (ILMD/Fiocruz Amazonia) and several hospitals are collaborating with the group on the project.
In addition to the new diagnostic methodology, the project calls for an investigation into the mechanisms involved in the blood clotting disorders associated with COVID-19, including platelet alterations. The principal investigator for this part of the project is José Carlos Nicolau, a professor at USP. The ongoing project described in the paper is also supported by FAPESP via grants awarded to Ester Sabino, also a professor at USP, and to Wagner José Fávaro and Fabio Trindade Maranhão Costa, both of whom are professors at UNICAMP.
The article “COVID-19 automated diagnosis and risk assessment through metabolomics and machine learning” can be retrieved from www.medrxiv.org/content/10.1101/2020.07.24.20161828v1.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





