

Discovery based on analysis of tissue and saliva samples from oral cancer patients shows a correlation between a signature comprising three peptides and the presence of lymph node metastasis (Gauging of the abundance of selected proteins in independent tissue sample using immunohistochemistry with antibodies/ Nature Communications)
Discovery based on analysis of tissue and saliva samples from oral cancer patients shows a correlation between a signature comprising three peptides and the presence of lymph node metastasis.
Discovery based on analysis of tissue and saliva samples from oral cancer patients shows a correlation between a signature comprising three peptides and the presence of lymph node metastasis.
Discovery based on analysis of tissue and saliva samples from oral cancer patients shows a correlation between a signature comprising three peptides and the presence of lymph node metastasis (Gauging of the abundance of selected proteins in independent tissue sample using immunohistochemistry with antibodies/ Nature Communications)
By Maria Fernanda Ziegler | Agência FAPESP – Researchers in Brazil have identified a correlation between oral cancer progression and the abundance of certain proteins present in tumor tissue and saliva. The discovery offers a parameter for predicting progression of the disease – whether cervical lymph node metastasis is present, for example – and points to a strategy for overcoming the limitations of clinical and imaging exams. It could also help guide the choice of an ideal treatment for each patient.
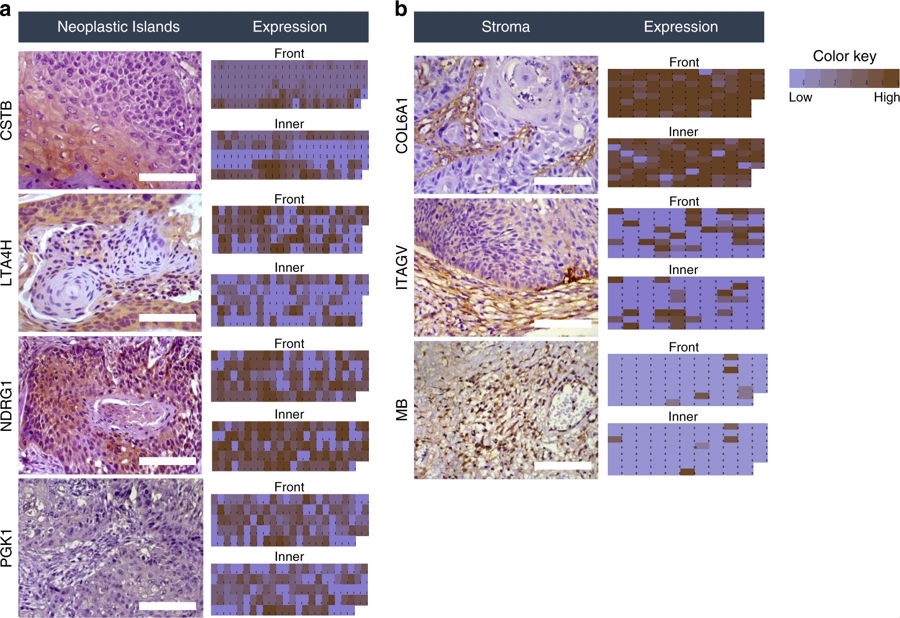
The study began in the discovery phase with a proteomic analysis of tissue from different tumor areas using 120 microdissected samples. In the verification phase, prognostic signatures were confirmed in approximately 800 tissue samples by immunohistochemistry and in 120 samples by targeted proteomics.
The study was supported by FAPESP and conducted at the National Energy and Materials Research Center (CNPEM) in partnership with the São Paulo State Cancer Institute (ICESP), the University of Campinas’s Piracicaba Dental School (FOP-UNICAMP), the Institute of Computing from the same university, the University of São Paulo’s Mathematics and Computer Science Institute (ICMC) in São Carlos, and the Dental School of the West Paraná University (UNIOESTE), in addition to other institutions in Brazil and abroad.
“The data led to robust results that are highly promising as guides to defining the severity of the disease. We suggested potential markers of the disease in the first phase of the study and verified these markers in the second phase, enhancing the reliability of the findings and showing that these markers are effective in classifying patients with cervical lymph node metastasis,” said Adriana Franco Paes Leme, a researcher at CNPEM’s National Bioscience Laboratory (LNBio) and the corresponding author of the article published in Nature Communications.
Mouth cancer, also known as oral squamous cell carcinoma (OSCC), is the most common type of malignant head and neck tumor. Prevalence and mortality are high, with some 300,000 new cases diagnosed per year worldwide and 145,000 deaths. Although it is relatively easy to detect, typically when a dentist notices an oral lesion, the disease is usually diagnosed when it is already at an advanced stage.
“We worked on the study for five years until we achieved this breakthrough,” Paes Leme told Agência FAPESP. “It was divided into two phases. In the first, we used discovery proteomics to identify and quantify tumor tissue proteins. The second phase of the study consisted of analyses using immunohistochemistry and targeted proteomics, for when you know precisely which proteins you want to quantify.”
Proteomics focuses on the identification, localization and functional analysis of the proteins in a sample, which may consist of tissue or cells, for example. The proteins are quantified, post-translational modifications are detected, and their activity and interactions are assessed.
Bioinformatics
In the first phase, the researchers used laser microdissection and proteomics to map the proteins in mouth cancer tissue and correlate them with the clinical characteristics of patients. This analysis permitted the identification of several proteins, such as CSTB, NDRG1, LTA4H, PGK1, COL6A1, ITGAV and MB, with differing levels of abundance depending on tumor area and links to key clinical outcomes.
In the second phase, after identifying and quantifying proteins in the 120-odd tumor tissue samples, the researchers deployed two protein verification strategies.
“One strategy consisted of gauging the abundance of the selected proteins in independent tissue samples using immunohistochemistry with antibodies. The other consisted of monitoring the same preselected targets in patients’ saliva,” Paes Leme said.
Saliva was chosen because this cancer is located in the mouth, where proteins could be secreted by neoplastic cells. “Saliva is a promising source of markers, as well as being a fluid obtained by noninvasive collection,” she explained. “We verified the proteins in saliva from 40 patients. Technical triplicates were analyzed to achieve the highest possible confidence level for the results in this phase of the study.”
After analyzing the saliva samples, the researchers used bioinformatics and machine learning techniques to arrive at prognostic signatures, verifying which of the proteins or peptides selected in the first phase could distinguish between patients with and without cervical lymph node metastasis. “In addition, we had valuable information about the clinical evolution of the patients who took part in the study as volunteers by donating samples of their saliva,” Paes Leme said.
From this result, it was possible to identify three specific peptides – LTA4H, COL6A1 and CSTB – that can be used as a signature to classify patients with and without cervical lymph node metastasis, offering the potential to help doctors overcome the limitations of clinical exams and guide personalized treatment strategies.
The scientists are now working on a new study designed to use translational techniques to build affordable biosensors capable of detecting prognostic signatures in patients’ saliva. Peptides currently have to be identified and quantified by mass spectrometry and proteomics, which are costly techniques and not often available in clinics and hospitals.
“We want to develop a simpler and cheaper method that can easily be used by health professionals to assess the progression of the disease on the basis of tests that can be performed in a dentist’s or doctor’s office, or in clinical labs. In the study we’ve just published, we were able to identify this prognostic signature by mass spectrometry. We now plan to develop a biosensor with a focus on the use of this signature so that it can be adopted for clinical use and help guide treatment decisions,” Paes Leme said.
The article “Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer” (doi:10.1038/s41467-018-05696-2) by Carolina Moretto Carnielli, Carolina Carneiro Soares Macedo, Tatiane De Rossi, Daniela Campos Granato, César Rivera, Romênia Ramos Domingues, Bianca Alves Pauletti, Sami Yokoo, Henry Heberle, Ariane Fidelis Busso-Lopes, Nilva Karla Cervigne, Iris Sawazaki-Calone, Gabriela Vaz Meirelles, Fábio Albuquerque Marchi, Guilherme Pimentel Telles, Rosane Minghim, Ana Carolina Prado Ribeiro, Thaís Bianca Brandão, Gilberto de Castro Jr, Wilfredo Alejandro González-Arriagada, Alexandre Gomes, Fabio Penteado, Alan Roger Santos-Silva, Márcio Ajudarte Lopes, Priscila Campioni Rodrigues, Elias Sundquist, Tuula Salo, Sabrina Daniela da Silva, Moulay A. Alaoui-Jamali, Edgard Graner, Jay W. Fox, Ricardo Della Coletta and Adriana Franco Paes Leme can be read at nature.com/articles/s41467-018-05696-2.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





