
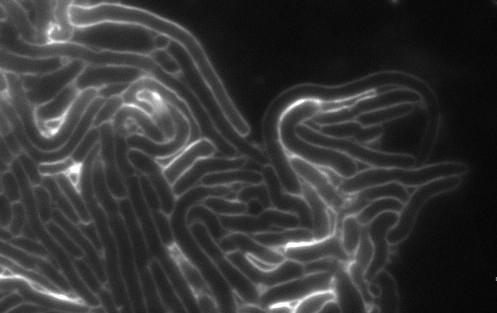
Toxic protein inhibits cell wall synthesis in rival bacteria. Discovery helps explain how pathogens cause imbalances in established microbial communities and could pave the way to the development of novel anti-microbial compounds (fluorescence image of bacteria attacked by bacterial toxin / Ethel Bayer-Santos)
Toxic protein inhibits cell wall synthesis in rival bacteria. Discovery helps explain how pathogens cause imbalances in established microbial communities and could pave the way to the development of novel anti-microbial compounds.
Toxic protein inhibits cell wall synthesis in rival bacteria. Discovery helps explain how pathogens cause imbalances in established microbial communities and could pave the way to the development of novel anti-microbial compounds.

Toxic protein inhibits cell wall synthesis in rival bacteria. Discovery helps explain how pathogens cause imbalances in established microbial communities and could pave the way to the development of novel anti-microbial compounds (fluorescence image of bacteria attacked by bacterial toxin / Ethel Bayer-Santos)
By André Julião | Agência FAPESP – Researchers at the University of São Paulo (USP) in Brazil have characterized a novel family of anti-bacterial toxins present in bacteria, including Salmonella enterica. This species uses toxic proteins to kill other bacteria in gut microbiota and facilitate colonization of the infected host’s gut.
The study is published in Cell Reports and featured on the cover of the journal.
The novel family’s founding member is the protein Tlde1 (type VI L,D-transpeptidase effector 1), which attacks bacterial cell wall precursors. It is secreted via the type VI secretion system or T6SS. Targeted bacteria continue growing but because their cell walls are weakened they eventually die as cell contents leak owing to osmotic pressure and lysis.
“This family of toxins has a hitherto undescribed mechanism. While other anti-bacterial toxins secreted by the same system destroy the already formed cell walls of target bacteria, this one acts on precursors so that they’re weak or cannot form at all,” said Ethel Bayer-Santos, a researcher in the University of São Paulo’s Biomedical Sciences Institute (ICB-USP) and principal investigator for the project, which is supported by FAPESP.
In a previous study, a research group that included Bayer-Santos described another secretion system (T4SS) in opportunistic bacteria of the species Stenotrophomonas maltophilia. They also described a toxin that inhibits bacterial growth (read more at: agencia.fapesp.br/32157).
“Bacteria selected these toxins during evolution and have been using them for thousands of years, so these discoveries point to therapeutic targets. Bacterial toxins may always have biotechnological potential and become anti-bacterial compounds in the future,” Bayer-Santos said.
Search
In their search for the protein Tlde1, the scientists analyzed the genome of Salmonella in the region neighboring the genes that encode the structural proteins in the secretion system (T6SS). They found a pair of genes with traits that signaled a toxic protein and another that conferred immunity. Anti-bacterial toxins are usually located in bacterial genomes near proteins that contain antidotes to the toxins. These antidotes are required to protect the microorganisms from their own weapons.
To test the function of this gene pair, the researchers first expressed a gene they thought was probably responsible for the toxic protein in a susceptible bacterium, Escherichia coli. It survived when the probable toxic gene was expressed in cytoplasm but died when expression occurred in periplasm, suggesting the toxin targets some structure in this part of the cell wall.
Next, they expressed in E. coli’s periplasm both the toxic protein and the probable immunity protein. This co-expression neutralized the toxic effect and the bacteria survived, confirming that the proteins in question are indeed a toxin and an immune protein respectively.
In evolutionary terms, T6SS is related to the apparatus of bacteriophages, viruses that infect bacteria. It comprises 13 structural proteins that are assembled into a weapon resembling a spear or harpoon with a sharp tip inside a retractable cytoplasmic sheath. The attacking cell ejects the harpoon full of toxic proteins from the sheath into the target cell.
Bioinformatics analysis showed that members of the Tlde1 family are present in several species of bacterium and that the family probably evolved from bacterial enzymes with a key role in cell wall synthesis. The next step in the project is an effort to understand by structural biology how an enzyme that had this role has ended up doing the opposite.
The first authors of the published paper are Stephanie Sibinelli-Sousa and Julia Takuno Hespanhol, who received scientific initiation scholarships (respectively grant no. 18/13819-1 and grant no. 18/25316-4). Sibinelli-Sousa is currently researching for a master’s degree at ICB-USP with FAPESP’s support.
The other authors are Gianlucca Gonçalves Nicastro, Robson Francisco de Souza and Cristiane Rodrigues Guzzo Carvalho, all of whom are affiliated with ICB-USP; Bruno Yasui Matsuyama, affiliated with the same university’s Chemistry Institute (IQ-USP); and Stephane Mesnage and Ankur Patel of Sheffield University in the UK.
The article “A family of T6SS antibacterial effectors related to l,d-transpeptidases targets the peptidoglycan” can be read at: www.cell.com/cell-reports/fulltext/S2211-1247(20)30794-4.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





