
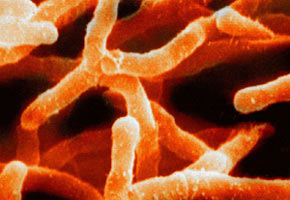
The object of the Unicamp study, Actinobacteria are important producers of secondary metabolites, molecules that could give rise to several types of drugs.
In the hope of discovering molecules with the potential to be used as immunosuppressive, antibacterial or anticancer drugs, Universidade Estadual de Campinas (Unicamp) researchers have dedicated themselves to studying a group of soil microorganisms known as Actinobacteria.
In the hope of discovering molecules with the potential to be used as immunosuppressive, antibacterial or anticancer drugs, Universidade Estadual de Campinas (Unicamp) researchers have dedicated themselves to studying a group of soil microorganisms known as Actinobacteria.

The object of the Unicamp study, Actinobacteria are important producers of secondary metabolites, molecules that could give rise to several types of drugs.
By Karina Toledo
Agência FAPESP – In the hope of discovering molecules with the potential to be used as immunosuppressive, antibacterial or anticancer drugs, Universidade Estadual de Campinas (Unicamp) researchers have dedicated themselves to studying a group of soil microorganisms known as Actinobacteria.
The study, titled “Exploring Brazilian biodiversity to obtain “non-natural” natural products,” was coordinated by Luciana Gonzaga de Oliveira, professor of the Chemistry Institute and funded in part through the FAPESP under the auspices of the Young Investigators in Emerging Centers Program.
Oliveira explains that the bacteria and fungi found in the soil are the greatest producers of molecules known as secondary metabolites.
Despite what their name might suggest, these molecules are essential for the survival of the microorganisms because they are involved in growth, development and reproduction. Furthermore, many secondary metabolites have bioactive properties, which researchers hope may make them effective drugs.
“The majority of metabolites with therapeutic potential were discovered in the golden age for antibiotics – between 1944 and 1972 – which gave rise to the medicines we use today, like erythromycin and rapamycin,” explains the researcher.
However, research dwindled as it became more difficult to find promising new molecules. Since then, few antibiotics have been developed – the vast majority of antibiotics used today are variations on active ingredients found more than 50 years ago. Pathogenic bacteria, on the other hand, have begun to develop resistance to several classes of existing drugs.
Hope for reversing this gloomy scenario has come with advances in inexpensive genetic mapping techniques, according to Oliveira. Many teams have invested in the complete sequencing of several Actinobacteria genomes, which revealed genes associated with the production of secondary metabolites.
“[We] realized that the biosynthetic potential of these strains has been underestimated. These microorganisms are a veritable machine for producing metabolites that can be explored. With the information from the genome, it is possible to predict the structures of these metabolite and in doing so estimate the microorganism’s potential for synthesizing new targets of interest,” he explained.
The team coordinated by Oliveira is particularly interested in two types of metabolites, reduced polyketides and nonribosomal peptides, which have numerous pharmacological activities. These molecules are synthesized by proteins and enzymatic complexes known as polyketide synthases (PKS) and nonribosomal peptide synthesis (NRPS), respectively.
“Research teams around the world have invested in studies guided by the genome for the identification of new metabolites. In Brazil, we have a rich diversity in terms of fauna, flora and also microorganisms, but we have few groups focused on research of this nature, which could boost the discovery of new drugs,” Oliveira said.
Sequencing
Instead of sequencing the entire genome of the Actinobacteria studied – which is not currently feasible due to cost – the Unicamp group is mapping the genetic fragments responsible for the production of PKS and NRPS enzymes. In this manner, it is possible to estimate the biosynthetic potential of several Actinobacteria strains, which includes the Streptomyces.
Initially, the researchers investigated 15 strains of bacteria and later expanded their studies to 80 strains. To concentrate their efforts on the most promising organisms, the researchers applied other tests that helped identify the presence of PKSs and NRPSs, selecting two other strains for further study.
“The studies involved identifying both the metabolites produced in cultures in laboratories and those that are not produced under these conditions, many times because the gene expression is not being activated in culture,” explains Oliveira.
Oliveira stresses that the entire project would have been facilitated and forecasts would have been made more rapidly if it had been possible to completely sequence the genomes of these microorganisms.
“In the near future, we want to sequence these two strains. This information would offer greater results and allow notable progress in this research area, making the country more competitive internationally,” he said.
Oliveira explains that the estimated cost of complete genome sequencing in a microorganism like these Actinobacteria is approximately R$ 30,000. “We are trying to work with other groups that are also interested in this area to invest in sequencing several microorganisms representing Brazil’s biodiversity,” she said.
To date, the project has involved the participation of a master’s student and six undergraduate students in the scientific initiation program. Partial results were presented in several national and international meetings, including the Brazilian Chemistry Society and the Ibero-American Symposium on Organic Chemistry.
Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.




